Lab 1. Invertebrate dissection
- Oyster dissection:
- o Place oyster with cup down and hinge facing you- use a shucker and a rock to open it- run the shucker along the edge and cut the adductor muscle.
- o Labial palps- direct food into the mouth or direct pseudofeces to a holding space next to the mantle, where it is expelled. The palps are a sorting organ for filter feeding.
- o Gonads- eggs vs. sperm- look like you would expect.
- o Heart- 2 auricles (dark red) and one ventricle (pale pink)
- Shrimp dissection:
- o Spot prawn?
- o Compound eye- many lenses!
- o Testis is dorsal and orange (chromatophores)
Lab 2. Invertebrate Histology
- Hepatopancreas + GIT
- Flexibacter in a finfish- gill lamellae + mucous
- Abalone- digestive gland, posterior esophagus, foot muscle, oocytes
- Abalone- withering syndrome- Rickettsia in a digestive gland
- Abalone hemocytes
- Clam- Bonamia in hemocytes
- Scallop- large RLOs in connective tissue and within digestive gland
- Scallop- Perkinsus quaswaldi in connective tissue
7/25/2012
Lab 3. Armina Dissection
- Armina eat sea pens- many slugs will swarm a sea pen at once and devour it. They also smell really fishy, even when they are alive.
- We dissected an adult Armina nudibranch, Accession # 12-1-6: we took three 2-3mm cross-sections of the caudal body and placed them in a tissue cassette, and covered them with a clean biopsy sponge. The cassette was placed in Invertebrate Davidson's fixative for 24 hours.
- We also took another, adjacent section and placed it in a 1-mL tube with 1 mL of 100% sterile, non-denatured ethanol.
- We made a squash preparation of adjacent tissue which is representative of our sample, to ensure that the correct tissue (digestive gland) was sampled. This tissue was identified microscopically as gonad, however.
- Morphometrics of our Armina: Total Length: 41.0 mm. Total Weight: 4.3 grams
- Gross observations: the nudibranch has 4 lesions on the dorsal skin- three similar, orange, 3x3mm lesions; and one linear, orange, 19x5mm lesion.
- Accession # / Total Length (mm) / Total Weight (g)
- 12-1-1 / 42.0 / 4.7
- 12-1-2 / 56.5 / 5.5
- 12-1-3 / 40.0 / 5.6
- 12-1-4 / 36.7 / 3.2
- 12-1-5 / 36.4 / 4.3
- 12-1-6 / 41.0 / 4.3

7/26/2012
Lab 4. DNA Extraction
Materials list:
Tissue samples in 95% ethanol
Qiagen Stool kit
Pipettes
Sterilized microcentrifuge tubes
Marking pens
70ºC incubator
Centrifuge
Vortex
Scalpels, blades, and tweezers
Weigh boats
Trays
Bleach bottles
Ethanol jar
Method: DNA Extraction (Qiagen Stool Kit)
- All the microcentrifuge tubes were pre-labeled for this procedure. The final 1.5 mL microcentrifuge tube was labeled with the following information: sample accession #, my initials, and the date. The sample # was placed on both the top and the side of the tube. We ran a group BLANK extraction as a negative control.
- Remove tissue from ethanol using good sterile technique and cut off a small piece of tissue approximately the size of ½ of a pencil eraser. The tissue was weighed: 0.1 g. The tissue was minced into small pieces and placed into a 2 mL microcentrifuge tube. Care was taken not to cross-contaminate samples.
- 1.4 mL Buffer ASL was added to each sample by adding 700 µl of Buffer ASL, vortexing for a minute and then adding another 700 µl of Buffer ASL. Once all ASL was added,the sample was vortexed continuously for 1 min until the sample was thoroughly homogenized to ensure maximum DNA concentration.
- The sample was heated for 5 min at 70º C.
- The sample was vortexed for 15 s and centrifuge sample at full speed for 1 min to pellet tissue particles.
- 1.2 mL of the supernatant was pipetted into a new 2 mL microcentrifuge tube and the pellet was discarded.
- 1 InhibitEX tablet was added to the sample and it was vortexed immediately and continuously for 1 min, until the tablet was completely dissolved. The sample was incubated for 1 min at room temperature to allow inhibitors to absorb the InhibitEx matrix.
- Sample was centrifuged at full speed for 3 min to pellet inhibitors bound to InhibitEX.
- All the supernatant was pipetted into a new 1.5 mL microcentrifuge tube and the pellet was discarded. Sample was centrifuged at full speed for 3 min. *in this step, the transfer of small quantities of pelleted material will not affect the procedure*
- 15 ul Proteinase K was pipetted into a new 1.5 mL microcentrifuge tube.
- 200 ul supernatant from step 9 was pipetted into the 1.5 mL microcentrifuge tube containing Proteinase K.
- 200 µl Buffer AL was added to the sample and it was vortexed for 15s. *DID NOT ADD Proteinase K directly to Buffer AL* It is essential that the sample and Buffer AL are thoroughly mixed to form a homogenous solution.
- Sample was incubated at 70 ºC for 10 min.
- Sample was removed from the incubator and briefly centrifuged. 200 µl of 95% molecular grade ethanol was added to the sample and it was vortexed for 15 seconds. The tube was briefly centrifuged.
- This mixture was carefully applied to a QIAamp spin column. If a white precipitate has formed, make sure to add this to the column. Did not wet the rim of the spin column (this can allow cross-contamination of samples in the centrifuge). Sample was centrifuged at 8000 rpm for 1 minute.
- The QIAamp spin column was placed in a clean 2 mL collection tube.
- The QIAamp spin column was carefully opened and 500 µl of buffer AW1 was added without wetting the rim. The sample was centrifuged at 8000 rpm for 1 minute.
- The 2 mL collection tube containing the buffer was discarded and place the spin column into a new collection tube.
- The spin column was opened and 500 µl of buffer AW2 was added without wetting the rim. Sample was centrifuged at full speed for 3 minutes.
- The spin column was placed into your final microcentrifuge tube, 100 µl of buffer AE was added to the column and sample was allowed to incubate at RT for 5 minutes.
- Sample was centrifuged at 8000 rpm for 1 minute
- The spin column was removed and thrown away.
PCR Lab:
For this lab we used molecular (PCR) techniques to test for RLOs in Armina californica. We first necropsied sea slugs and sampled digestive gland tissue for PCR and histology. Next, we extracted DNA from the digestive gland of each of the sea slugs sampled. Using this extracted DNA as template, we set up a conventional PCR (cPCR) using 3 sets of primers: (1) universal bacterial - EUB A/B, (2) Ehrlichia - EHR16s, and (3) WS-RLO specific - RA 3-6/RA 5-1. The RA primers amplify a 160 bp fragment of the 16S WS-RLO genome.
We set up multiple cPCR reactions / master mixes for the primer sets to be tested.
Supplies:
Fwd Primer
Rev Primer
DNA samples
Positive control
Sterile PCR water
PCR tubes
Sterile 1.5 mL microcentrifuge tubes
Methods
- We calculated the volumes needed for the PCR reactions (see master mix recipes below). We made at least 10% more than we needed, so that we did not run out of master mix. All calculations were be recorded as follows below.
- Wipe down bench with 10% bleach.
- To a 1.5 mL tube, all reagents except the DNA template were added.
- Tube was vortexed briefly (pulse).
- PCR tubes were labeled with the sample # and primers. Two tubes were labelled (–) for the negative control and two (+) for the positive control.
- For the WS-RLO recipe, 18 μL of master mix was added to each of the PCR tubes.
- For the generic recipe, 23 μL of master mix was added to each of the PCR tubes.
- 2 µL of PCR water was added to each negative control.
- 2 μL of template DNA was added to each tube, taking care not to cross contaminate between samples.
- 2 µL of known positive template was added to the positive control tubes.
- The microtubes were vortexed briefly (pulse) and spun down.
- The thermal profiles were run for cPCR.
| Reagents |
uL/reaction |
| Immomix 2x |
12.5 |
| BSA (10mg/ml) |
1.5 |
| Fwd primer (10um) |
0.8 |
| Rev primer (10uM) |
0.8 |
| sH20 |
7.4 |
| Template |
2 |
| Total |
25 |
BSA (10mg/ml): 1.5 x 8 = 12 uL/rxn
Fwd primer (10 um): 0.8 x 8 = 6.4 uL/rxn
Rev primer (10 um): 0.8 x 8 = 6.4 uL/rxn
Sterile water: 7.4 x 8 = 59.2 uL/rxn
Template (extracted sample): 2 uL/rxn (do not multiply by 8)
Total = 23 x 8 = 184
| Thermal profile |
Temp |
Time |
| Step1 |
95 |
10min |
| 45 cycles of |
||
| Step 2 |
95 |
15 sec |
| Step 3 |
60 |
1min |
WS-RLO cPCR recipe
PCR primers from Andree et al. 2000 with optimized protocol of Friedman et al. 2008
| [Stock]Beginning concentration |
[End]Solution Concentration |
Per |
|||||
| Reagent |
reaction (ml) |
||||||
| 5 X Buffer |
5x |
1 |
X |
4 |
|||
| MgCl2 |
25 mM |
1.5 |
mM |
1.2 |
|||
| BSA |
10 mg/ml |
400 |
ng/ml |
0.8 |
|||
| H2O |
11.08 |
||||||
| dNTP's |
10 mM |
200 |
µM |
0.4 |
|||
| RA 3-6 |
100 pmol/ml |
0.5 |
µM |
0.1 |
|||
| RA 5-1 |
100 pmol/ml |
0.5 |
µM |
0.1 |
|||
| Taq |
5 U/ml |
1.6 |
U |
0.32 |
|||
| Template |
2 |
||||||
| Total Reaction Volume |
20 ml |
||||||
5X Buffer: 4x8 = 24 uL/rxn
MgCl2: 1.2x8 = 9.6 uL/rxn
BSA: 0.8x8 = 6.4 uL/rxn
Sterile water: 11.08x8 = 88.64 uL/rxn
dNTP's: 0.4x8 = 3.2 uL/rxn
RA 3-6: 0.1x8 = 0.8 uL/rxn
RA 5-1: 0.1x8 = 0.8 uL/rxn
Taq: 0.32x8 = 2.56 uL/rxn
Template: 2 uL/rxn (do not multiply)
Thermal profile
| Time |
Temp (°C) |
|
| Step 1 |
3 min |
95 |
| Step 2 |
1 min |
95 |
| Step 3 |
30 sec |
62 |
| Step 4 |
30 sec |
72 |
| Repeat steps 2-4, 40 times |
||
| Step 5 |
10 min |
72 |
Andree, K.B., C.S. Friedman, J. D. Moore, and R. P. Hedrick. 2000. A polymerase chain reaction assay for the detection of genomic DNA of a Rickettsiales-like prokaryote associated with withering syndrome in California abalone. Journal of Shellfish Research. 19(1): 213-218.
Order of PCR products in microtubes:
1. Negative control
2. Blank sample template
3. 12-1-6 CSC sample
4. 12-1-6 GS sample
5. 7-29-10 C-Dead AP sample
6. Positive control
7/27/2012
Materials list:
Pipettes
100 bp Ladder
Loading dye
Agarose
200 ml flask
Graduated cyclinder
Microwave
1X TBE
Gel box & combs
Power supply
UV transilluminator
Ethidium bromide
Lab Mat
Methods
- 1.5 g agarose was weighed and added to a 200 ml flask
- 100 mL 1 X TBE (diluted from 10X TBE; 100 ml 10X TBE + 900 ml RO water; (V1) (C1) = (V2)(C2)) was added to the flask
- The solution was brought to a gentle boil in the microwave for ~ 3 minutes (1.5min, stop, gently swirl, 1.5min)
- 10 µL of ethidium bromide was added (A POTENT MUTAGEN!!), the solution was swirled gently, allowed to cool for a few minutes and poured into the gel mold
- Gel combs were placed in and gel was allowed to set for ~ 15min
- 1X TBE was added to the gel box (~ ½” over the top of the gel)
- 7 µl of 100 bp molecular weight ladder was pipetted in the far end wells
- 5 µl of loading dye was added to the PCR products *UNLESS dye is already incorporated into your master mix buffer. Change tips for every sample to avoid contamination.
- 7 µl of your PCR product + loading dye was pipetted into each well.
- Gels were run at 115V for 45 mins or until dye is ¾ way down the gel.
- Gel was carefully removed and examined under UV light (wearing nitrile gloves and lab coat).
- Gel was photographed.
72712_Gel.xlsx
Gel Visualization:
The only PCR product that was positive by gel resolution was the 12-1-6 CSC sample, using the universal bacterial primer. All other fluorescing bands were in the wrong location or were postive due to primer dimers.
7/30/2012
Laby Collection Trip:
We went to False Bay and did three shallow 10m transects, within which a minimum of 100 eelgrass blades were counted and measured, and the presumed Labyrinthula zosteraeinfections were recorded and quantified. We also evaluated 4 quadrants within each transect, in which we quantified the density of eelgrass plants. On each transect, we checked every eelgrass shoot within approx. 6” of the tape. In very dense areas we only checked eelgrass directly under the tape. Each shoot will be sized (into 5 categories) and its severity of infection recorded. Severity of infection will be on a 4 point scale: 0 (healthy), 1 (1 small lesion), 2 (2-3 moderate lesions), 3 (over 50% of blade affected, and/or more than 1 blade in a shoot affected).
Materials:
For field work we will need:
3 transect tapes
2 plexiglass sheets for mapping
temperature, pH meter
data sheets, clipboards, pencils
plastic bags for samples

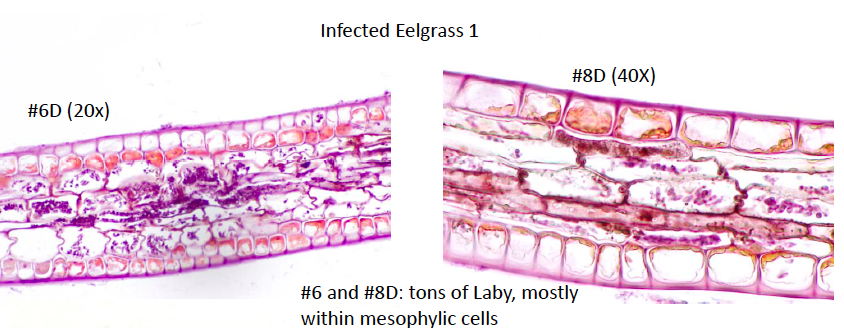
Laby Histology:
We microscopically evaluated the infected eelgrass samples that were collected last week, and classified the Laby infections according to number of Laby cells per HPF.
| SLIDE # |
# Laby cells/ 40x field |
||||
|---|---|---|---|---|---|
| PC2-D3 |
1 |
2 |
0 |
7 |
1 |
| 2 |
3 |
0 |
2 |
1 |
|
| 3 |
0 |
0 |
1 |
0 |
|
| 4 |
1 |
0 |
0 |
0 |
|
| 5 |
1 |
0 |
0 |
0 |
|
| PC2-D13 |
1 |
1 |
0 |
0 |
0 |
| 2 |
0 |
0 |
0 |
0 |
|
| 3 |
0 |
0 |
1 |
0 |
|
| 4 |
0 |
0 |
0 |
0 |
|
| 5 |
1 |
0 |
0 |
0 |
|
| PC2-D15 |
1 |
2 |
0 |
||
| 2 |
3 |
2 |
|||
| 3 |
1 |
2 |
|||
| 4 |
0 |
0 |
|||
| 5 |
3 |
2 |
|||
Laby Culture:
Labyrinthula zosterae isolates can be cultured either in solid or liquid media. Recipes for both are below. Cultures need to be transferred every week or two depending on room temperature for solid media and about 2-5 days for liquid media. All cultures will be grown at room temperature and exposed to light during the day.
We placed presumed infected eelgrass samples on culture plates with Laby media, using sterile technique and after briefly immersing the eelgrass sample in 70% alcohol.
Modified SSA Agar
- Filter sterilize at least 1 L of seawater using to 0.45um or smaller
- Check salinity using a refractometer and adjust to 25ppt using Nanopure water (or DI water). Adjust slowly, mix well, and check salinity using refractometer frequently.
- Add 1 L of 25ppt filtered seawater to a 2 L flask. Add an autoclavable stir bar and the following:
- "USB Nobel Agar": 12g
- Germanium dioxide: 1.5 mg
- Yeast extract: 0.1g
- Peptone: 0.1g
- Glucose: 1.0g
- Loosely cover with foil and autoclave for 20 minutes on the liquid cycle.
- While media is in the autoclave heat up water bath to 37 oC and thaw 10 mL aliquot of Horse Serum and 25 mL 100X Pen/Strep.
- Once autoclaved, temper the media for 30 minutes to between 50 and 55 oC after mixing gently - avoid creating bubbles in the agar.
- Add thawed 10 mL horse serum and 25 mL 100X Pen/Strep.
- Mix or stir adequately, making sure not to create any bubbles and pour the plates.
- Let agar cool and then store at 4 oC until use.
Modified SSA Broth
1. Filter sterilize at least 1 L of seawater using to 0.45um or smaller
2. Check salinity using a refractometer and adjust to 25ppt using Nanopure or DI water. Adjust slowly, mix well, and check salinity using refractometer frequently.
3. Add 1 L of 25ppt filtered seawater to a 2 L flask. Add an autoclavable stir bar and the following:
- Germanium dioxide: 1.5 mg
- Yeast extract: 0.1g
- Peptone: 0.1g
- Glucose: 1.0g
4. Split into four 250 mL corning jars (the ones with the orange lids) with lids loose and covered in foil.
5. While media is in the autoclave heat up water bath to 37 oC and thaw 10 mL aliquot of Horse Serum and 25 mL Pen/Strep.
6. Once autoclaved, swirl media to mix well and cool the media for 30 minutes to between 50 and 55 oC.
7. Add thawed 2.5 mL horse serum and 6.25 mL 100X Pen/Strep to each jar
8. Store at 4°C in clean room fridge until use.
7/31/2012
Laby Collection Trip:
We went to Beachcomber Bay on Orcas Island and evaluated three shallow 10m transects, within which a minimum of 100 eelgrass blades were counted and measured, and the presumed Labyrinthula zosterae infections were recorded and quantified. We also evaluated 4-5 quadrants within each transect, in which we quantified the density of eelgrass plants. Within the quadrants, we also quantified the numbers of adult snails and number of snail eggs masses, as well as the number of flowering eelgrass plants.
We plated 6 diseases eelgrass samples on the media listed above, for culture.
8/1/2012
Laby Collection Trip:
We went to Picnic Cove on Shaw Island and evaluated 2 deep and 3 shallow 10m transects, within which a minimum of 100 eelgrass blades were counted and measured, and the presumed Labyrinthula zosterae infections were recorded and quantified. We also evaluated 4-5 quadrants within each transect, in which we quantified the density of eelgrass plants.
DNA Extractions
Extraction & PCR of laby DNA—An adjacent slice of blade tot he sections taken for histology will be preserved in 95% ethanol for subsequent PCR (labeled, screw-cap vial filled with 95% non-denatured ethanol).
For this lab we used a commercially available kit which relies on the silica membrane extraction, called the Qiagen ® Stool kit.
Materials list:
Tissue samples in 95% ethanol
Qiagen Stool kit
Pipettes
Sterilized microcentrifuge tubes
Marking pens
70ºC incubator
Centrifuge
Vortex
Scalpels, blades, and tweezers
Weigh boats
Trays
Bleach bottles
Ethanol jar
Method: DNA Extraction (Qiagen Stool Kit)
- All the microcentrifuge tubes needed for this procedure were minimally pre-labeled. You will need a final 1.5 mL microcentrifuge tube, please label this with the following information: your sample accession #, your initials, and the date. Please put the sample # on both the top and the side of the tube. Remember to always run a BLANK extraction as a negative control. My sample is Picnic Cove Healthy-12 (PC-H-12), preserved in 95% ethanol.
- Removed blade from ethanol using good sterile technique and cut a small piece of laby-infected blade approximately the length of a pencil eraser (try to minimize the amount of eelgrass – grasses contain pigments which could potentially inhibit subsequent PCR reactions). The sample was weighed and recorded. Sample was minced into small pieces and placed into a 2 mL microcentrifuge tube, taking care not to cross-contaminate samples.
- 1.4 mL Buffer ASL was added to each sample by adding 700 µl of Buffer ASL, vortexing for a minute and then adding another 700 µl of Buffer ASL. Once all ASL was added, solution was vortexed continuously for 1 min until the sample was thoroughly homogenized. Please note: It is important to vortex the samples thoroughly as it will insure maximum DNA concentration.
- Solution was heated for 5 min at 70º C.
- Solution was vortexed for 15 s and centrifuge sample at full speed for 1 min to pellet blade particles.
- 1.2 mL of the supernatant was pipetted into a new 2 mL microcentrifuge tube and discard the pellet.
- 1 InhibitEX tablet was added to each sample and vortexed immediately and continuously for 1 min until the tablet was completely dissolved. Solution was incubated for 1 min at room temperature to allow inhibitors to absorb the InhibitEx matrix.
- Sample was centrifuged at full speed for 3 min to pellet inhibitors bound to InhibitEX.
- All the supernatant was pipetted into a new 1.5 mL microcentrifuge tube and the pellet was discarded. The sample was centrifuged at full speed for 3 min. *in this step, the transfer of small quantities of pelleted material will not affect the procedure*
- 15 ul Proteinase K was pipetted into a new 1.5 mL microcentrifuge tube.
- 200 ul supernatant from step 9 was pipetted into the 1.5 mL microcentrifuge tube containing Proteinase K.
- 200 µl Buffer AL was added, and vortexed for 15s. *DO NOT ADD Proteinase K directly to Buffer AL* It is essential that the sample and Buffer AL are thoroughly mixed to form a homogenous solution.
- Solution was incubated at 70 ºC for 10 min.
- Samples were removed from the incubator and briefly centrifuge. Add 200 µl of 95% molecular grade ethanol to the sample and vortex for 15 seconds. Briefly centrifuge the tubes.
- This mixture was carefully applied to a QIAamp spin column. If a white precipitate has formed, make sure to add this to the column. Do not wet the rim of the spin column (this can allow cross-contamination of samples in the centrifuge). Centrifuge at 8000 rpm for 1 minute.
- The QIAamp spin column was placed in a clean 2 mL collection tube.
- The QIAamp spin column was carefully opened, and 500 µl of buffer AW1 was added without wetting the rim. Centrifuge at 8000 rpm for 1 minute.
- The 2 mL collection tube containing the buffer was discarded, and the spin column was placed into a new collection tube.
- The spin column was opened and 500 µl of buffer AW2 was added without wetting the rim. Centrifuge at full speed for 3 minutes.
- The spin column was placed into the final microcentrifuge tube, 100 µl of buffer AE was added to the column and allowed to incubate at RT for 5 minutes.
- Tube was centrifuged at 8000 rpm for 1 minute.
- The spin column was removed and thrown away, leaving a tube of DNA.
8/2/2012: Laby PCR
Polymerase Chain Reaction (PCR)
Primer Design:
Using Geneious software, information was gathered from examining the L. zosterae 154 sequence (west coast strain; GenBank: AF265335.1) against the L. zosterae MBL 93-2 sequence (east coast strain; GenBank: AF265334.1). An alignment of the 2 sequences were conducted and new primers were designed in Geneious using Primer 3 in a non-conflicting region (200-1180 bp) for the primers to lay down. Below is the new primer information:
Forward Primer:
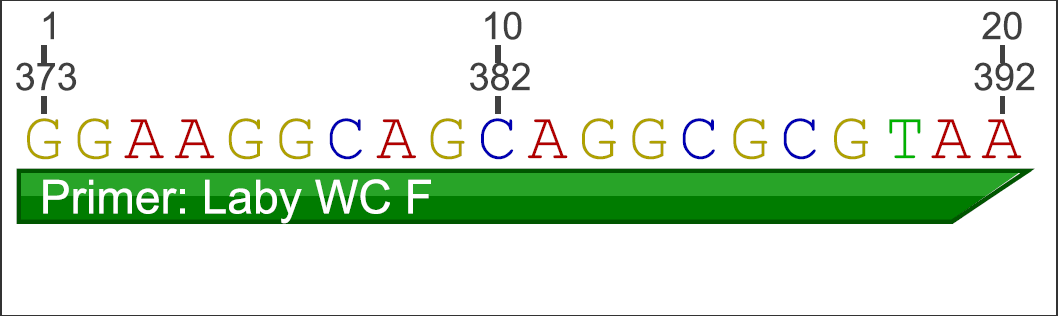
Reverse Primer:

conventional PCR (cPCR):
- The total number of reactions was calculated, making sure to account for a positive control and no template negative control.
- Using the recipe below, we calculated the amount of each ingredient to add to the master mix.
| Ingredient |
Amount per Rxn |
# of Rxns X 1.1* |
To add to Master Mix |
||
| GoTaq Mix |
12.5 uL |
X |
10 |
= |
137.5 |
| BSA |
1.5 uL |
X |
10 |
= |
16.5 |
| Forward Primer |
0.8 uL |
X |
10 |
= |
8.8 |
| Reverse Primer |
0.8 uL |
X |
10 |
= |
8.8 |
| PCR H2O |
7.4 uL |
X |
10 |
= |
81.4 |
- Using sterile technique, all ingredients were added to a microcentrifuge tube that can hold the total volume.
- The solution was briefly vortexed and spun down.
- 23 uL master mix was aliquoted into each tube or well.
- 2 uL template was added to each tube, making sure to keep track of what sample is where.
- The solution was capped, briefly vortexed, and spun down.
- Samples were placed in the thermal cycler and run using the following thermal profile:
| Time |
Temp (°C) |
|
| 1. |
10 min |
95 |
| 2. |
30 sec |
95 |
| 3. |
30 sec |
50 |
| 4. |
60 sec |
72 |
| 5. |
Repeat steps 2-4, 40 times |
|
| 6. |
10 min |
72 |
| 7. |
Hold @ 4 deg C |
| 1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
| Neg control |
PC-H-12 |
PC-1-D12 |
PD-1 |
Pd-2 |
LD-1 |
LD-2 |
LH |
PH |
Pos control |
8/3/2012: Gel Electrophoresis
Gel Electrophoresis:
1. 1.5 g agarose was weighed and added to 200 ml flask
2. 100 mL 1 X TBE was added to the flask
3.The solution was brought to a gentle boil in the microwave for ~ 3 minutes (1.5min, stop, gently swirl, 1.5min)
4. 10 µL of SybrSafe was added to the solution, swirled gently, allowed to cool for a few minutes and poured into the gel mold
5. Combs were placed in and gel was allowed to set for ~ 15min
6. 1X TBE was added to the gel box (~ ½” over the top of the gel)
7. 7 µl of 100 bp molecular weight ladder was pipetted in the far end well
8. 5 µl of loading dye was added to this.
8/6/2012
Today we learned how to measure water alkalinity (measures the ability of the water to buffer acid) using the DL15 titrator- see handout for details.
We also toured the Ocean Acidification Lab to follow up a great lecture.
8/7/2012- Vibrio tubiashii in Pacific Oysters
We plated the 6 different strains of Vibrio tubiashii on 6 T1N2 (clear) plates, along with a streak of Aeromonas media to allow us to evaluate the probiotic properties of A. media on Vibrio. The 6 strains of Vibrio were plated on a TCBS (green) plate. We used sterile technique (flamed loop between each streak). Our group used the Vibrio strain RE98. The plates were allowed to incubate at room temperature for 24 hours.
We learned how to count oyster larvae microscopically. Larval concentration is calculated according to this equation: V1C1 = V2C2 ==> x (Larval conc) = 40 larvae (150 ul).
We also had a demonstration of how to use the spectrophotometer to determine water pH.
photo courtesy of Jenna Malek
8/8/2012
Question 1: Are all strains equally pathogenic?
We will conduct the following experiment:
We will challenge larval oysters with the bacterial strains sent to us to assess their pathogenicity using a LD50 (lethal dose to 50% mortality) assay in which oysters will be bath-exposed to multiple ten-fold serial dilutions of each bacterium grown at ambient or reduced pH. We will monitor oysters for signs of morbidity or mortality over a 48-hour period. We will attempt to re-isolate the bacteria from our experiment and then conduct some preliminary phenotypic characterization on any bacteria recovered. These characterizations should give us a tentative identification of the bacteria.
Day 1: Oyster challenges comparing bacterial strains
Serial dilutions of bacteria
Plate bacteria
Challenge oysters with serial dilution of bacteria at ambient pH
Return at days 1 and 2 post-inoculation to count plates and assess oyster mortality
For this exercise, we began by creating a serial dilution of the stock bacterial culture for the LD50 challenges.
Step 1: Serial dilutions and spread plates
For disease challenge:Stock culture containing bacterial isolate
2 - 12-well tissue culture plates with C. gigas larvae
14 culture tubes with SW for dilutions
1000 ul pipets and pipet tips
100 ul pipets and pipet tips
6 T1N2 plates
2 TCBS plates
Hockey stick
Ethanol and sand bath
Bunsen burner
Parafilm
Bacterial loop
32 x 250 ml beakers
For total alkalinity and spectrophotometric pH:
Parafilm
HCl for alkalinity
m-cresol purple for spec pH
CRMs for TA
pH standards for NBS and spec pH
TA machine
Ocean optics machine
Cuvettes
50ml serological pipettes and pipettor
Ziploc bags
25°C Water bath
Lens paper
Kimwipes
1. The Vt and Am isolates were obtained for the challenge were are conducting. Our Vt isolate is RE22-Ambient.
2. A serial dilution of the Vt to 10^-7 was made
- Started by adding 9 mL of seawater to 7 culture tubes.
- Each of the culture tubes was labeled with one of the following dilution labels (10^-1, 10^-2, 10^-3, 10^-4, 10^-5, 10^-6, 10^-7).
- Dilution series was started by taking 1 mL of bacterial suspension and adding it to the tube labeled 10-1. This is the first 10-fold dilution. It was mixed well making sure to hold cap with thumb securely.
- 1 mL of your 10^-1 dilution tube was added to the tube labeled 10^-2. This is the second dilution. It was mixed well.
- The above process was repeated until the dilutions were finished out to 10^-7.
3. A serial dilution of Am to 10^-7 was made using the methods in Step 2.
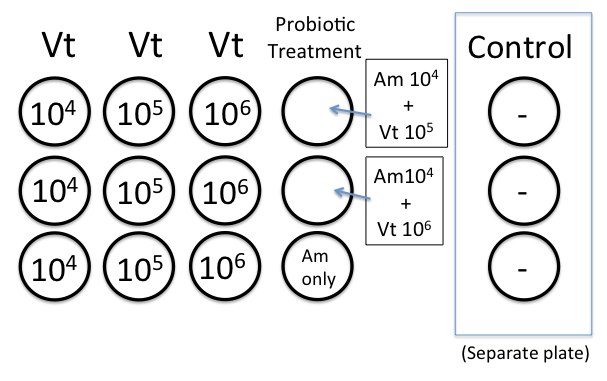
4. 6 T1N2 plates were labeled with a dilution (10^-5 through 10^-7) and bacterial strain (Am or Vt). Dilution, the date, and initials on the were recorded on the BOTTOM of each petri plate. Obtain 2 TCBS plates and label 10^-6.
5. 0.1 mL of bacterial suspension was plated onto the appropriate T1N2 plate, starting from the lowest dilution (10^-7) to the highest dilution (10^-5), and using the same pipet tip.
6. 0.1 mL culture was plated onto a TCBS plate, using one plate for each culture. Only the 10-6 dilutions were plated. TCBS (thiosulfate citrate bile salt sucrose) agar is a selective media for different types of bacteria of the genus Vibrio. Different species of Vibrio produce different colored colonies on the TCBS agar. The color change is indicative of...
7. Hockey stick was dipped into ethanol sand bath and flamed. The hockey stick was cooled by touching it to a sterile, dry area of the plate and then the bacterial suspension was spread in a circular motion. Again, we started with the lowest dilution to highest dilution for all.
8. Plates were left on the bench right-side up for about an hour. Plates were sealed in parafilm, inverted, and incubated at 30C overnight.

Step 2: Oyster challenge
Serial dilutions of bacteria from Step 1Larval oysters
LD50 challenge with V. tubiashii
Seawater chemistry was assessed prior to and after the experiment (spec pH and TA- see spectrophotmetirc pH and Total alkalinity protocols). For this we prepared triplicate 250 ml beakers containing 200mls of
- seawater at right CO2 level
- seawater at right CO2 level with 10^5 of Vt/ml and 2,000 C gigas larvae
- seawater at right CO2 level with 10^5 of Am/ml and 2,000 C gigas larvae
- Seawater samples:
- Seawater and larvae were screened into a clean beaker to remove larvae.
- pH of all three types seawater samples (a-c above) was taken with the TA machine’s pH probe.
- The spec pH cuvette was overfilled with sample type (a)=seawater only, cap and processed as per spec pH protocol.
- Remaining seawater was used to measure alkalinity as per alkalinity protocol.
- Wells were pre-filled with 24-100 uls of larvae at a density of ~40 larval oysters/well with 3 replicates for each Vt dilution.
- Wells were inoculated with 100 ul of the appropriate bacterial dilution (10^4, 10^5, or 10^6) to bring total well volume to 4mls. Did not inoculate the control wells with Vt.
A. media after 24h incubation at 37oC = ~1.5x10^8 CFU/ml
V. tubiashii after 24h incubation at room temp (22oC) = ~3.0x10^9 CFU/ml
We will add 100ul A. media culture to each well in the probiotic test. --> 3.0x10^9 x 0.1ml = 3.0x10^8 CFU/ml
3.0x10^8 CFU/ml (100ul of culture)= 7.5x10^7 CFU/ml using the original culture
4.0x10^0 mlSW (volume in the well)
Original Vt culture = 7.5x10^7 CFU/ml
(-1) 1:10 dilution = 7.5x10^6 CFU/ml (too high)
(-2) 1:10 dilution = 7.5x10^5 CFU/ml
(-3) 1:10 dilution = 7.5x10^4 CFU.ml (good)
Original A. media culture: 1.5x10^8 CFU/ml x 0.1 ml = 1.5x10^7 CFU/ml
1.5x10^7 CFU/ml = 3.75x10^6 CFU/ml
4.0x10^0 mlSW
(-1) 1:10 dilution = 3.75x10^5 CFU/ml
(-2) 1:10 dilution = 3.75x10^4 CFU/ml (we will use this one)
- On day 1 post-challenge
- Oyster mortality in each well was recorded. We only counted dead larvae; total larvae will be tallied at the end of the trial.
- Bacteria were re-isolated on T1N2 and TCBS plates from one control and one 10-6 well using the streak plate method. We were sure to include treatment, dilution, well # sampled, the date, and initials on the bottom of the petri plates.
- T1N2 dilution plates (10-5 through 10-7) that we made yesterday were counted, and colony growth, morphology, and color were examined for both T1N2 and TCBS plates.
- On day 2 post-challenge
- The number of dead oysters in each well was recorded.
- All larvae in each well were killed using a small amount of diluted bleach and total amount of larvae in each well was recorded.
- Ended experiment and cleaned up.
- NOTE: Our experiment will be maintained at the same pH the Vt culture was grown in. We made sure that the plates went into the appropriate area (either low pH or ambient environment).
8/9/2012
DAY 2: Bacterial Characterization and Quantify Larval Mortality
1: Count spread plates and calculate concentration of starting culture
A plate with 30 to 300 colonies was chosen to count. We counted the colonies and multiplied this number by the dilution. When the bacteria was plated, we only added 0.1 mL of the bacterial suspension to the plate so we had to account for that dilution (which is 1/10).CFU/mL = # colonies * dilution (e.g. 105) * plate dilution (10)
We recorded the plate counts and calculations as well as the shape, elevation and color of the colonies (e.g. circular, raised, off-white) for both your T1N2 and TCBS plates.
2: Recover bacteria and make a streak plate.
Sterile technique was used for this step.
We made 2 streak plates, one on T1N2 and one on TCBS for each: control, 10-2, and 10-6 for both ambient and low pH wells. A streak plate is an important technique for not only re-isolating the bacteria but also obtaining pure cultures. The goal of a streak plate is to produce single bacterial colonies; as one progresses from the first quadrant to the second, the amount of bacteria is diluted. This technique is outlined below:
Supplies
Oyster sampleInoculating loop
70% ethanol + sand
Burner
3 T1N2 plate
3 TCBS plate
METHODS
- A sterile petri dish was divided into four sections using a pen (labeled the underside of the plate).
- Metal loop was flamed until red. Loop was cooled before use by touching an unused area of plate that is still sterile.
- Loop was dipped into the well of the oysters. Carefully lifted lid of agar plate, keeping lid over plate. Streaked back and forth in 1st quadrant without going over same agar twice. Put inoculating loop into 70% ethanol and sand.
- Loop was flamed as before, then touched agar and streaked through 1st quadrant 2-3 times and then moved to 2nd quadrant and streak without going over same area twice. Loop was placed in 70% ethanol and sand.
- Loop was flamed again and streaked through the 2nd quadrant, moving the streak to 3rd. Loop was flamed and repeated until the 4th quadrant is streaked. Loop was placed in 70% ethanol and sand.
- Loop was flamed BEFORE putting away or setting down.
- The plates were parafilmed, to be examined next week.
3: Analysis and bacterial identification
- Gram stain on bacteria:
- Selected a colony from plate and suspended in a drop of water on a glass slide.
- Let air dry
- Heat fixed slide by briefly passing over a flame
- On a rack, flooded slide with crystal violet for 1 min
- Washed briefly in tap water to remove excess crystal violet
- Flooded with Lugol’s or Gram’s iodine 1 min
- Washed briefly in tap water
- Immediately, rapidly de-colorized slide with alcohol-acetone solution until the stain ran past the lower edge of the section
- Washed slide immediately in tap water
- If the section appears too blue repeat steps h and i
- Counterstained slide with safranin ~15 sec
- Bloted dry, added immersion oil and viewed at 100x
Examine results:
Gram positive bacteria.........................................purple/dark blue/black
Filaments of nocardia and mycobacteria.............dark blue but may have red sheathGram negative bacteria…………………...........redNuclei ..................................................................red
Serum Agglutination Test
- Added 50 uL of sterile seawater and 25 uL of the culture to one slide (control)
- To one slide (test slide) added 25 uL of sterile seawater and 25 uL of culture and 25uL of the thawed polyclonal antibody.
- Gently mixed solutions on both slides with sterile pipet tips
- Incubated at room temperature for 5 mins
- Viewed at 10x or 4x on the scope to compare degree of agglutination of the cells
After disease challenge is completed and we have tallied the larvae in each well, we used the mortality counts for the 24-hr and 48-hr time points to determine proportion of larval mortality at each bacterial concentration. Averaged the proportions before entering the numbers into the calculator. (Calculations based on Saganuwan 2011)
*See excel spreadsheet with LD50 calculator to calculate LD50 for each treatment.*
- Median survival =
- Average Vt dose causing 50% mortality =
- Log(answer (b.))
- Answer (a.) * Answer (c.)
- Antilog of answer (d.) provides you with LD50 dose of Vt.
24 hr post inoculation ambient 24 hr post inoculation HIGH
Fill out shaded areas only EXAMPLE DATA
Vt Concentration Resulting in Below 50% Mortality Log Vt Dose Average Proportion Larval Survival (0-1 value) % Survival Vt Concentration Resulting in Below 50% Mortality Log Vt Dose Average Proportion Larval Survival (0-1 value) % Survival
18500000 7.26717172840301 0.632 63.2 100000 5 0.93 93
Vt Concentration Resulting in Above 50% Mortality Vt Concentration Resulting in Above 50% Mortality
25000000 7.39794000867204 0.237 23.7 1000000 6 0.4 40
Survival 18500000 Survival 100000
Median: 0.665822784810127 25000000 Median: 0.188679245283019 1000000
Dose: 1.35135135135135 Dose: 10
Log(dose) 0.130768280269024 Log(dose) 1
0.087068500533552 0.188679245283019
Antilog of: 7.35424022893657 Antilog of: 5.18867924528302
LD50: 2.26E+07 LD50: 1.54E+05
8/11/2012:Cleaned up around the lab and joined groups for our projects.
8/13/2012: Today we learned how to use NCBI. We completed an exercise called My Favorite Gene.
Bioinformatics lab (from Sonia's notebook):
Downloaded the SwissProt database onto the lab computers (CL_14 à Users à Shared à EIMD_blast à db) and unzipped it. The database was then created through the terminal:
%% cd /Applications/blast/bin
%% ./makeblastdb -in [input file in .fasta] -out [output file name, no .fasta] -dbtype prot
Input: /Users/Shared/EIMD_blast/db/uniprot_sprot.fasta
Output: /Users/Shared/EIMD_blast/db/uniprot_sprot
- Note that the Applications folder used is also under CL_14, NOT under fhl_guest.
- On a Mac, directory can be set by dragging the appropriate folder from Finder into the Terminal.
%% ./blastx -query [input file in .fasta] -db [uniprot_sprot, no .fasta]
- Query: /Users/Shared/EIMD_blast/query/QPX_transcriptome_v1.fasta
- Database (db): /Users/Shared/EIMD_blast/db/uniprot_sprot
First opened file (/Users/Shared/EIMD_blast/query/QPX_transcriptome_v1.fasta) in TextWrangler. (Say “No” to any options that come up when TextWrangler opens.) To prevent misreading of the sequence names/sequences, we did a reformat:
- Find: 1 (space) Contig (space)
- Replace all with: 1_Contig_
- Resave file.
In Terminal:
%% ./blastx
-query [input file]
-db [database file, no .fasta]
-evalue 1e-20
-max_target_seqs 1
-outfmt 6
-out [outfile]
-num_threads 2
Notes:
*query used: /Users/Shared/EIMD_blast/query/QPX_transcriptome_v1.fasta
*db: /Users/Shared/EIMD_blast/db/uniprot_sprot
*out: /Users/Shared/EIMD_blast/out/qpx_transcr_swisspro.txt
*E-value of e^(-20) => only HIGHLY similar sequences will be selected
*max_target_seqs – only return the first (highest) hit for each contig
*outfmt 6 = tab-delimited
*num_threads = number of CPUs to use
Note that the command above should be written in a single line, with spaces instead of enters/tabs. I’ve written it out that way to make it easier to read all the arguments.
=
Let the program run on the terminal overnight. Final location of the resulting text file:https://catalyst.uw.edu/sharespaces/download/17360/318727?html=1&url=https://catalyst.uw.edu/sharespaces/download/17360/318727
8/14/2012Today we learned about CLC command functions and BLASTed the 2 QPX libraries.
Notes:Use CLC to do de novo assembly on the two QPX libraries.When you map the reads back to the contigs on the backbone, set settings at 2 and 5.
RPKM = reads per kilobase of exon model per million mapped reads- normalizes for library size
In the “experiment” the two tables are put together
Next BLAST the transcriptome in Galaxy – enter the e-value to -20
 We also completed and presented our My Favorite Gene projects.https://docs.google.com/document/d/1e8tkWwtvleAMruHLnuCKTQ3tb3F0I9Cf7mZMjjS6tU4/edit
We also completed and presented our My Favorite Gene projects.https://docs.google.com/document/d/1e8tkWwtvleAMruHLnuCKTQ3tb3F0I9Cf7mZMjjS6tU4/editWe worked in Galaxy to enter the QPX data and get an output of the gene identities, which will allow us to evaluate protein function after we put it all together.
The limit on our BLAST table was E-25- gives us a lot of confidence in our hits.
- Joined Awesome BLAST file (column 4) with the SPID and Description (column 1). Used "Join 2 Datasets" in "Join, Subtract, Group".
- Want the e-value to be low; the e-value tells you how much confidence you have in your gene identity.
- Bit score is another way to look at similarity, represents how good an alignment is.
>Next I joined the QPX Annotated Transcriptome (SP), column 4 with Associations Uni SwissPro, column 2
>3 types of ontologies: biological process, molecular structure, and cellular component. We are interested in the Biological Process genes.
>Imported the GoSlim term> takes every annotated GO term and generalizes it.
>Next I joined GOSlim.txt (column 1) to the previously joined files (GO files, column 24), to scale down the information and make it easier to handle. Generalizes the groupings and gives us less overall info.
>At this point we have about 18,000 lines of data.
>Download this file as a tab-delimited file, to then open it in Excel- open the file in Excel.
>Can then delete files you don't want, and keep the following columns: QPX Contig #, e-value, gene name, Go term, Go Slim term, and gene ontology domain.
>Then make a Pivot table in Excel, under Data. Row labels- put Go Slim. Also put Go Slim under values. Then make a pie chart out of the pivot table.
From Sonia's Lab Notebook:
Bioinformatics lab:
Joining tables in Galaxy:
Galaxy’s setup is:
- Tools on the left
- Files and dialogue boxes in the middle
- History (steps taken, numbered in reverse chronological order) on the right. Every step is recorded, and steps that change or create new data tables are saved in the box of that step as new tables.
- “Eye” allows you to view the result of that step
- “Pencil” allows you to edit attributes of the file
- “X” cancels the step or deletes the table
- If you click on the title in the box, it will give you a “save” button that you can use to download the data table.
- Log into Galaxy (https://main.g2.bx.psu.edu/)
- Upload file(s) to Galaxy:
- Left bar: Get data
- Upload file from your computer (here, data = the qpx mapped reads)
- Check upload: Click on the “eye” symbol in the green box with the file name on the right-hand side.
- Separate different identifiers in Column 3 (column 2 in non-headed version):
- Left bar: Text manipulations
- Convert delimiters to tab
- In the dialogue box, make sure “pipes” are being converted.
- Check: Click on the “eye” in the right-hand green box labeled “Convert on data”. Can rename file if desired by clicking the “pencil”.
- Import previously-established history of SwissPro accession numbers and gene names:
- Go to https://main.g2.bx.psu.edu/u/sr320/h/databases.
- Click on the “eye” under “SPID and description”
- Click “upload” (green circle with the cross)
- Join two tables:
- MAKE SURE THE ROWS IN THE COLUMNS TO JOIN ARE IDENTICAL. (That is, that elements in the desired column in one table EXACTLY match the rows in the desired column in the other table.)
- Left bar: Join, subtract and group
- Join two datasets
- Here: In the dialogue box join “Convert on data” (or equivalent), c4 (c3 for non-headed version); with “SPID and description”, c1.
- Keep all lines and fill unjoined rows.
- Check: Click on the “eye” next to the “Join” box on the right.
- Add protein functions:
- Import the “SPID and GO #” database from the above address.
- Join two data tables as above, merging the columns with the SwissPro ID numbers.
- Check result. GO data should be in column 24 (23 in unheaded data table)
- Import the “GO and GO_slim” database from the above address.
- Join the data tables by GO number.
- Check result.
Gene Enrichment Analyses- focus on temperature differences. See statistically accurately the physiologic differences in the pathogen at difference temperatures.
Used limited short-read data, did de novo assembly to get 11,000 contigs, annotated, and classified with GO Slim. Then did reference mapping.
>Average contig coverage- takes average vertical coverage. Then found novel sequences (all).
>Used RNA Seq to evaluate differentially expressed genes- gene enrichment analyses > take a subset f genes and see if there's a statistical difference between the genes and the backbone. The differentially expressed gene set is a subset of the total transcriptome.
>Subset may be statistically significant in the subset but may not be significant in the whole transcriptome. To see if there is over- or underrepresentation of certain GO terms.
>Identify these significantly different GO terms, put them in a software.
For QPX, we found 684 differentially expressed genes, looked to see if any GO terms were over/underrepresented- to see if the 2 clam populations were different in terms of immunity. This allows us to see what specific genes are responsible for which actions. We look for increased translation of proteins to maintain homeostasis. We specifically looked at genes with a 2-fold difference in expression at varying temperatures.
>We found 4895 genes with a -2-fold expression, meaning they have decreased expression at 21oC.
>We found 112 genes with a +2-fold expression, meaning they have an increased expression at 21oC.
>We deleted all genes that were not 2-fold expressions, to give us a list of differentially expressed genes.
Our enrichment analyses will be based on the SwissPro ID number. Thus, we compared the SwissPro IDs from the entire transcriptome and the SwissPro IDs from the differentially expressed genes (DEGs).
>We then joined the QPX experimental unique reads of DEGs (c1) to the Awesome BLAST results (c2), and named it Enrichment Analysis Based on SwissPro ID 2.
>Then opened this file in Excel.
>We then used david.abcc.ncifcrf.gov to look at the over/underrepresented genes between the two data sets. This process is called gene set enrichment analysis.
>In David, first put the DEGs into David under Uniprot ID; submit this to gene conversion too, then Convert All under the gene list.
>Next put the entire transcriptome as the background data, under Uniprot accession number.
>then have to tell David whether the results are genes or background, and give each a name.
>Then go to Annotation Summary Results.
>We are now interested in Gene Ontology.
>Go to chart (Functional Annotation Chart), which tells you what biological processes are enriched (over/underrepresented).
>Can also look at pathways.
>Want a table for Biological Processes > FAT table. Open this up in Excel, have to remove the ~.
>Open up Revigo online, to make pictures out of our data. Revigo wants 2 columns- we entered the GO numbers in one column and the p-values in the second column.
Idea: make an x-y plot of the unique reads at 21oC and 10oC to look at differential gene expression.
In Summary:
- trimmed reads
- De novo assembly
- De novo assembly blast against Swiss-Prot (terminal) (provides just SP ID)
- De novo assembly used to map reads from each library (RNA-Seq)
- Used Galaxy to get much more information (gene description and GO information)
- Differentially expressed genes were identified (2-fold)
- Used Galaxy to get SP ID of just the Differentially expressed genes
- Enrichment Analysis using DAVID- using Background SP ID and DEG (Gene list) SP ID
- Downloaded files (GO FAT BP) from DAVID
- Tweaked GO FAT BP in Excel
- REVIGO used to visualize Enriched GO BP Processes
8/16/2012
- Abalone - Metagenomics Analysis
- Three Libraries - Libraries Total RNA Sequences
- Blasting __NCBI blast against nt__ (blast tables available using this link)
This will provide most thorough taxonomic representation (Galaxy)
- Targeted blasting to specific databases (ie VirusRefSeq, BacteriaRefSeq) (stand alone blast / terminal)
- Blasting between libraries (What’s different?)
__BlackAb_WSP_assembly..>__ 10-Aug-2012 06:51 529k
__BlackAb_WSO_assembly..>__ 09-Aug-2012 17:57 454k
__BlackAb_NWS_assembly..>__ 09-Aug-2012 17:01 456k
-ALSO you could map the raw (short) reads back to a particular genome, sequence etc. (Galaxy)
8/17/2012
Why Study genetic variation?
==>
- population level
- evolution
- conservation biology
- metagenomics
- gene variation -> resilience -> epigenetics
- pathogen variability
- predictions of future changes
- source pathogen
- phenotypic plasticity
How?
==>
- SNPs
- RFLPs -> DNA fingerprinting
- microsatellites